Family History Extraction using Clinical Narratives
Link to full Graduate Research Assistant
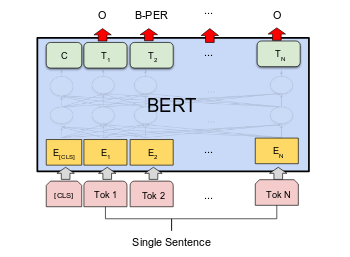
For my graduation project, I worked with Prof. Meliha Yetisgen as a graduate research assistant in the BioNLP lab and worked on a project that aimed at extracting family history from patients’ clinical narratives. The project required me to develop an end-to-end system from preparing the training dataset for the individual tasks to selecting the model, algorithm, and implementation.
This project can be divided into two main subtasks:
• Entity identification (family members and disease names) - This involves getting the family members and disease names (unassociated). I decided to use Clinical BERT for Named Entity Recognition by fine-tuning BERT for Token Classification. This method required to use of the transformer and add a final decision layer for NER. This method gave very promising results and was able to identify most, if not all entities.
• Family history extraction - the system is expected to associate family members and corresponding observations. The strategy for this subtask is to start small and build on top by analyzing the results. Most baseline relation extraction models use POS tags to denote a relation. Using the POS tags as a feature in the relation extraction task, I am using Clinical BERT for relation extraction as well. The implementation for this follows a ”matching the blanks” method. Since this is similar to masked language modeling, it would be a good way to use BERT for relation extraction.
The dataset provided for this task is from Patient Provided Information (PPI) questionnaires, which are usually stored in semi-structured/unstructured formats.